New Genetic 'Operating System' Facilitated Evolution of 'Bilateral' Animals
September 30, 2014
By Kim McDonald

Bilateral animals, such as this nudibranch, have three germ layers—ectoderm, mesoderm and endoderm—in the embryo, whereas more primitive radial animals, such sea anemones (below), possess only two germ layers—an ectoderm and endoderm.
Wikipedia Commons
The evolution of worms, insects, vertebrates and other "bilateral" animals—those with distinct left and right sides—from less complex creatures like jellyfish and sea anemones with "radial" symmetry may have been facilitated by the emergence of a completely new "operating system" for controlling genetic instructions in the cell.
That's the hypothesis of molecular biologists at UC San Diego. They report in the October 1 issue of the journal Genes & Development that this new system of controlling gene networks, analogous to a new computer operating system, paved the way for new animal body plans, just as different operating systems allow the development of new kinds of computer apps.
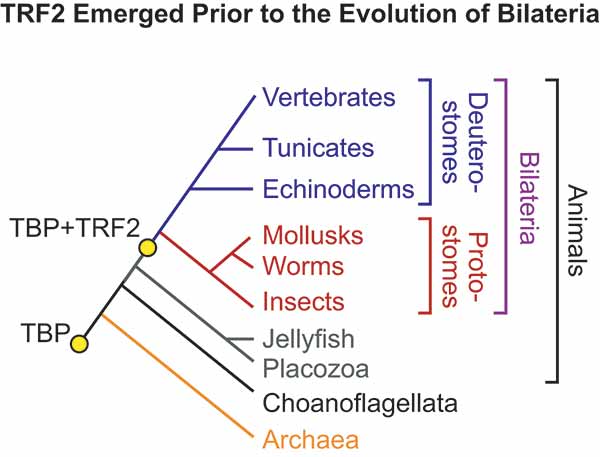
One key player in this theory is an ancient protein termed "TATA box-binding protein," or TBP, which is found in organisms ranging from archaebacteria to humans. A billion years ago, TBP served as the core of a single "operating system" for gene expression.
The UC San Diego scientists focused their attention on a more recent protein related to TBP termed "TBP-related protein 2," or TRF2. They found that TRF2 is present in bilateral animals, and is absent in animals that lack bilateral symmetry, such as jellyfish, sea anemones and sponges. This observation inspired the idea that the emergence of TRF2 provided animals with an entirely new operating system with new gene expression programs ("apps") that facilitated the evolution of bilateral organisms.

While their hypothesis initially seemed far-fetched, the UC San Diego scientists continued to analyze TRF2 in greater detail. They found that the new facts were like interlocking pieces of a jigsaw puzzle that fit together into a coherent picture. For example, bilateral animals have three germ layers—ectoderm, mesoderm and endoderm—in the embryo, whereas more primitive radial animals, such as jellyfish and sea anemones, possess only two germ layers—an ectoderm and endoderm.
"It turns out that TRF2 appears to be important for the formation of the mesoderm, the germ layer that is missing in radial organisms and is present in bilateral organisms," said Jim Kadonaga, a biology professor at UC San Diego who headed the study "In this manner, the pieces of this scientific jigsaw puzzle fell into place."
"The emergence of TRF2 essentially doubled the regulatory capacity of the organisms because TBP and TRF2 can function mostly independently of each other," he explained.
The scientists began studying TRF2 while conducting research on a related project, which they published this past June in Genes & Development. In that study, they reported the discovery that TRF2 plays a key role in the production of the 80 proteins that make up the ribosome—the protein factory of the cell.
"Those findings revealed that TRF2 can support the transcription of a specific network of genes independently of TBP, and suggested the existence of two operating systems based on TBP or TRF2," said Kadonaga.
Sascha Duttke, a graduate student in Kadonaga's laboratory trained in evolutionary biology, was curious as to how and why this second system of transcription evolved. So he examined the presence or absence of TRF2 among different animal groups with the help of Russell Doolittle, one of the world's experts on protein evolution and an emeritus professor of biology and chemistry at UC San Diego. This led to the finding that TRF2 is in bilaterians but not non-bilaterian animals.
"In our current model, there was originally only a single TBP-based operating system, and then the emergence of the new TRF2-based operating system led to new gene networks ('apps') that facilitated the emergence of bilateria," said Kadonaga. "These new gene networks included those that are involved in the development of the body plan and the mesoderm. We postulate that the new TRF2-based networks provided the extra diversity of regulatory function that led to the evolution of more complex organisms—specifically the bilateria, which constitute about 99 percent of living animals."
In addition to Duttke and Doolittle, the UC San Diego team included Yuan-Liang Wang, a biologist in Kadonaga's lab. Their study was funded by a grant from the National Institutes of Health's National Institute of General Medical Sciences (R01 GM041249).
