New multiplex labeling technique fills gap between in situ hybridization and microarray analysis
October 11, 2004
By: Kim McDonald
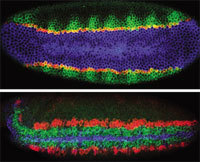
TRIPLE DELIGHT: Ventral (top) and ventro-lateral (bottom) views of Drosophila embryos triple-labeled to visualize expression patterns. The top image (early blastoderm) shows snail (blue), single minded (red), and rhomboid (green); the bottom image (early gastrulation) shows short gastrulation (blue), ventral nervous system defective (green), and intermediate nervous system defective (red). Credit:
http://superfly.ucsd.edu/
davek/gallery.html
Scientists at the University of California, San Diego, recently described a novel combination of new and preexisting technology that permits them to visualize five RNAs simultaneously in a single cell.1 The technique, called multiplex labeling, is a variant of multiplex fluorescent in situ hybridization (M-FISH), and was developed by Dave Kosman, a staff scientist in the laboratories of UCSD biologists Ethan Bier and William McGinnis.
Kosman and coworkers, in collaboration with scientists at Molecular Probes (a division of Invitrogen in Carlsbad, Calif.) used the method to directly detect microRNA in an intact Drosophila embryo, as well as nascent RNA transcripts in a nucleus at the site of transcription. "It's the first technique that I've seen where I was convinced that one could look at more than three gene products at a time in the same cell," says Scott Fraser, director of the Biological Imaging Center, California Institute of Technology. He notes that in the past researchers looked at complex cellular events by piecing together images from several embryos. Fraser is currently working with the paper's authors to implement the procedure in his own lab.
Multiplex labeling builds upon previous labeling techniques such as conventional in situ hybridization (ISH) and M-FISH, but allows more genes to be studied at higher resolution, says Kosman. In traditional ISH, RNA is labeled with an epitope tag such as digoxigenin, which is recognized by antibodies coupled to horseradish peroxidase or alkaline phosphatase to generate a colorimetric signal. But this method lacks multiplexing capability, a problem that can be overcome by using fluorescence-based detection.
Rather than using a single labeling method, Kosman and colleagues used a three-technique strategy to maximize gene coverage, says Greg Cox of Molecular Probes and an author on the paper. Fluorescently labeled secondary antibodies or tyramide signal amplification to boost sensitivity was used to detect rare transcripts and genes expressed at low-to-moderate levels. "Using the tyramide amplification system, we estimate this method to be about 10 times more sensitive than existing histochemical detection methods," says Bier.
Cox developed a two-step, direct RNA-labeling technique that could detect more abundant transcripts. In this method, aminoallyl-UTP is incorporated into the target RNA during probe synthesis. The aminoallyl linker serves as a docking site for the probe of choice: in this case, Molecular Probes' Alexa Fluor dyes, which are brighter and bleach less than conventional fluorescent probes, according to Cox. Kosman notes that this technique allows more efficient labeling than standard fluorescent labeling of nucleic acids, in which the UTP is modified by a bulkier fluorescent moiety that is more difficult to incorporate during probe synthesis.
SEEING SPOTS Kosman and colleagues also used fluorescent labeling to create a coding system that would identify nascent RNA transcripts, by taking advantage of the gene transcription sites' ability to be visualized as "nuclear dots" via FISH. Both individual fluorophores and multicolor dye combinations were used to expand the number of genes viewed simultaneously. "By using two of those dyes in combination and then overlaying the colors, you can actually make a third color, which expands the multiplex capability of the system greatly," Cox explains.
This combinatorial approach to fluorescence can theoretically detect expression of as many as 50 different genes in a single embryo, says Bier, who explains that multiplex labeling fills a gap between genome-wide expression analysis and ISH techniques that detect one or two genes at a time. "A big need, it seemed, was to fill this gap," he says, "to be able to look at high precision, like you can with in situ hybridization, but with far more than just one or two genes, and be able to unequivocally tell precisely what genes are on in a single cell at a particular time in an embryo in space and in time."
Geneticist Mike Levine at UC, Berkeley, is using multiplex labeling in an attempt to directly visualize long-range enhancer-promoter interactions in Drosophila embryos. In several cases, RNAs associated with an enhancer on one chromosome appeared to interact with those of a promoter on another chromosome, suggesting that the researchers were viewing the process of transvection. This visualization "is possible only with this method," says Levine, who further notes that his entire laboratory is now adopting Kosman's approach over conventional ISH. "The lab [people are] very set in their ways and reluctant to change methods. But they are all switching," Levine says.
Kosman emphasizes that his method is still a work in progress and that several "inconvenient" aspects of the protocol, including the use of antibodies to detect the probes, still need to be worked out. Currently, at most only five genes can be visualized simultaneously without combinatorial fluorescence; Kosman would like to see six or seven at a time.
--Aileen Constans
1. D. Kosman et al., "Multiplex detection of RNA expression in Drosophila embryos," Science, 305:846, Aug. 6, 2004.
