Scott Rifkin
Research
My laboratory studies how stochastic, genetic, and environmental variation is filtered through development and physiology to produce phenotypic variation and how evolutionary forces shape that process. We work primarily at the level of gene expression, taking advantage of technologies for visualizing gene expression dynamics at single-transcript resolution, among others. Current projects in the lab include probing the dynamics and robustness of the genetic networks underlying sex-determination and intestinal specification in nematodes and investigating how natural variation affects the dynamic properties of signaling and regulatory networks in yeasts.
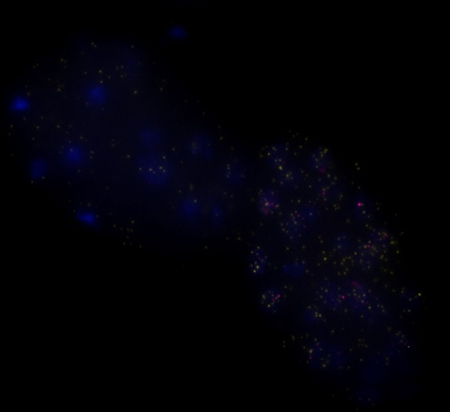
Individual transcripts of xol-1 (yellow) and sdc-2 (red) in early C. elegans embryos. Blue spots are DAPI-stained nuclei.
Select Publications
- L Du, S Tracy, SA Rifkin (2016). Mutagenesis of GATA motifs controlling the endoderm regulator elt-2 reveals distinct dominant and secondary cis-regulatory elements. Developmental Biology. (in press)
- M Maduro, G Broitman-Maduro, H Choi, F Carranza, AC-Y Wu, SA Rifkin (2016). MED GATA factors promote robust development of the elegans endoderm. Developmental Biology. 404:66-79.
- SR Stockwell, CR Landry, SA Rifkin (2015). The yeast galactose network as a quantitative model for cellular memory. Molecular Biosystems. 11: 28-37.
- AC-Y Wu, SA Rifkin (2015). Aro: A machine learning approach to identifying single molecules and estimating classification error in fluorescence microscopy images. BMC Bioinformatics. 16: 102.
- MA Bakowski, CA Desjardins, MG Smelkinson, TA Dunbar, IF Lopez-Moyado, SA Rifkin, CA Cuomo, ER Troemel (2014). Ubiquitin-mediated response to microsporidia and virus infection. in C. elegans. PLoS Pathogens. 10: e1004200.
- CR Landry*, SA Rifkin* (2012). The genotype-phenotype maps of systems biology and quantitative genetics: distinct and complementary. in Evolutionary Systems Biology. (Soyer, ed.). Advances in Experimental Medicine and Biology Series. 751:371-398. *equal contribution
- SA Rifkin (ed.) (2012). Quantitative Trait Loci: methods and protocols. Methods in Molecular Biology, 871. Springer, New York.
- SA Rifkin (2011). Identifying fluorescently labeled single molecules in image stacks using machine learning. in Molecular Methods for Evolutionary Genetics (Orgogozo & Rockman eds.) Methods in Molecular Biology. 772: 329-348.
- CR Landry, SA Rifkin (2010). Chromatin regulators shape the genotype–phenotype map. Molecular Systems Biology. 6:434.
- A Raj*, SA Rifkin*, E Andersen, A van Oudenaarden (2010). Variability in gene expression underlies partial penetrance in Caenorhabditis elegans. Nature. 463: 913-918. *equal contribution
- A Raj, P van den Bogaard, SA Rifkin, A van Oudenaarden, S Tyagi (2008). Imaging individual mRNA molecules using multiple singly labeled probes. Nature Methods 5:877-889.
- Y Gilad, SA Rifkin, JK Pritchard (2008). Revealing the architecture of gene regulation: the promise of eQTL studies. Trends in Genetics 24:408-413.
- CR Landry, B Lemos, SA Rifkin, WJ Dickinson, DL Hartl (2008). Genetic properties influencing the evolvability of gene expression. Science 317:118-121.
- Y Gilad, A Oshlack, SA Rifkin (2006). Natural selection on gene expression. Trends in Genetics 22:456-461.
- SA Rifkin, D Houle, J Kim, KP White (2005). A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature 438:220-223.
- Y Gilad, SA Rifkin, P Bertone, M Gerstein, KP White (2005). Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Research 15:674-680.
- SA Rifkin, J Kim, KP White (2003). Evolution of gene expression in the Drosophila melanogaster subgroup. Nature Genetics 33:138-144.
- SA Rifkin, J Kim (2002). Geometry of gene expression dynamics. Bioinformatics. 18:1176-1183.
- SA Rifkin, K Atteson, J Kim (2000). Constraint structure analysis of gene expression. Functional and Integrative Genomics. 1:174-185.
- KP White, SA Rifkin, P Hurban, DS Hogness (1999). Microarray analysis of Drosophila development during metamorphosis. Science. 286:2179-2184.
Biography
Scott Rifkin received his PhD from Yale University and was a postdoctoral fellow at Harvard and an NIH postdoctoral fellow at MIT. He joined the faculty of UCSD in the summer of 2009.

